
- The sequence B family – rivalling the gold standard DNA polymerases
- Speed vs accuracy: when proofreading is a problem
- Quit (uracil) stalling: overcoming the challenge of modified DNA for greater application possibilities
- Harnessing the power of the KOD variants: tailored enzymes to suit emergent technology-driven method
The recent explosion in new DNA amplification methods has been fortified by novel and tailored DNA polymerases with enhanced fidelity and a variety of improved features, including thermostability and resistance to inhibitors as found in crude sample targets. An expanding repertoire of polymerases suitable for DNA amplification have ensued – and KOD polymerases are increasingly finding their role within these applications. Going beyond their natural forms, customised KODs with distinct attributes enable R&D scientists to pursue new amplification methods powered by tailored polymerases. Technologies which are hinged on specialised, tailored DNA polymerase options compatible with challenging samples suitable for versatile applications.
The Sequence B family – Rivalling the Gold Standard DNA Polymerases
Over the last 60 years, extensive biochemical characterisation of numerous DNA polymerases has facilitated their use in a wide range of molecular biology and biotechnology applications. With such a variety of choice available, how can R&D scientists select appropriately?
Suitable for several PCR applications, thermostable Taq DNA polymerase is still considered by many to be the industry standard. However, the performance of Taq is limited in more challenging applications, such as those requiring high fidelity synthesis of long (> 2 kb) amplicons, and amplification of GC-rich sequences. Most notably, Taq DNA polymerase lacks proofreading activity, the determinant of fidelity; indeed, comparisons of base substitution error rates between non-proofreading (10-2 to > 10-6) and proofreading polymerases (10-6 to 10-7) are striking.
Enter the archaeal B-type DNA polymerases with integral 3´→5´ exonuclease activity for mis-paired base removal, contributing to the hallmark advantage of higher fidelity.
Among them, Pyrococcus furiosus (Pfu DNA polymerase) is nearly 4 times more stable than Taq DNA polymerase at 95°C; yet, along with most archaeal proofreading DNA polymerases, it exhibits limited processivity (< 20 bases) in vitro (Table 1).
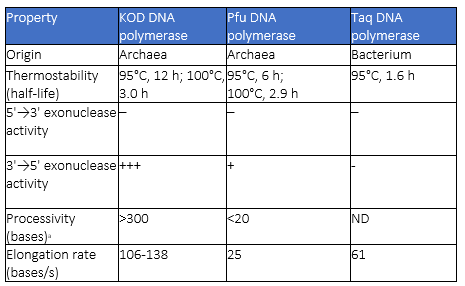
Table 1: A comparison of the properties of the KOD, Pfu and Taq DNA polymerases
a the average number of nucleotides incorporated before a disassociation event
However, there is one member of the family that lacks the processivity limitations of other archaeal DNA polymerases: the KOD DNA polymerase derived from Thermococcus kodakarensis.
Although KOD DNA polymerase's optimum temperature (75o C) and mutation frequency (3.5 x 10-3) is similar to those of Pfu DNA polymerase, it displays four distinct hallmark features over it:
- an elongation rate (100-130 nucleotides/s) that is ~5 times higher
- a processivity 10 to 15 times greater
- effective amplification from GC rich targets
- crude sample compatibility
Collectively, these characteristics result in minimised PCR run times, dropping it to under a third of the time required for Taq-mediated PCR. Combined with the elimination of sample preparation and sonication, even greater savings on laboratory time are achievable, alongside eliminating
potential sample loss incurred by purification.
The KOD DNA polymerase then has clear advantages – but it is not without its Achilles heel…
Speed vs Accuracy: When Proofreading is a Problem
While the presence of an integral 3´→5´ exonuclease (proofreading) activity is essential in PCR, this feature becomes a liability in applications that champion the need for speed. Higher accuracy and processivity come at the expense of elongation velocity when PCR goes the distance (>5 kbp).
The reason for the dip in product yield? The KOD DNA polymerase exonuclease domain, but the target length limitation resulting from hampered speed can be addressed.
One genetically engineered variant of the KOD DNA polymerase is the 3′→5′ exonuclease-deficient mutant, KOD exo(-) DNA Polymerase. The substitution of amino acid residues in conserved exonuclease motifs removes the exonuclease activity while maintaining effective amplification of GC rich target sequences taken from crude samples.
Available with HotStart functionality, KOD exo(-) DNA Polymerase can be applied to real-time PCR. As part of the KOD SYBR qPCR Master Mix for long target, GC-rich amplification from crude samples, it is also suitable for library quantification which can be applied to next generation sequencing.
Quit (Uracil) Stalling: Overcoming the Challenge of Modified DNA for Greater Application Possibilities
Despite exceptional processivity, KOD DNA polymerase poses a drawback – a "read-ahead" function that detects uracil (dU) residues in the template strand and stalls synthesis – unlike Taq. With dUTP a common strategy to prevent carry-over contamination resulting in DNA containing uracil, stalling of DNA synthesis opposite uracil compromises the performance of archaeal DNA polymerases at large.
The issue of uracil stalling is compounded in next generation sequencing (NGS) applications where it constitutes baseline noise and may interfere with detection of low-level gene mutations. Moreover, techniques requiring dUTP (i.e., dUTP/UDG decontamination methods) or uracil-containing oligonucleotides are also eliminated, limiting the range of applications of high-processivity and high-fidelity proofreading DNA polymerases.
However, this challenge can be overcome with (yet again) polymerase tailoring. Through genetic modification of the KOD DNA Polymerase dUTP binding pocket, the KOD DNA polymerase’s ability to bind uracil is eliminated, enabling KOD DNA polymerase to conduct effective amplification through uracil and other modified residues. This uracil tolerance compliments bisulfite-sequencing workflows where traditional high-fidelity DNA polymerases are incompatible.
Harnessing the Power of the KOD Variants: Tailored Enzymes to Suit Emergent Technology-Driven Methods
Although Taq and Pfu DNA polymerases have found a firm foothold in PCR applications, their limitations restrict their incorporation in more complex applications. New technologies are driving the need for improved DNA amplification protocols and the elaboration of new tailored methods – and as a key element, novel, faithful and processive DNA polymerases are a driving force in the field. KOD DNA polymerases are one powerful option, whose hallmarks of homogeneous amplification, high-fidelity, high-processivity, efficiency and sensitivity position them among the gold standards of polymerases – particularly for GC-rich targets. Better still, tailored variants offer a variety of improved features – enabling R&D scientists to embrace the changing landscape of application demands and overcome the challenge of modified bases and inhibitors that plague crude samples.
Find out more about KOD Multi & Epi DNA Polymerase, a uracil-tolerant KOD DNA polymerase suitable for multiplex PCR in NGS analyses. Discover how the KOD DNA Polymerase performs as part of the KOD One PCR Master Mix for amplification of NGS libraries and more
SEKISUI Diagnostics delivers KOD DNA polymerase optimised for your applications – from long and accurate PCR and genotyping to NGS analysis driven by emerging technologies – we empower R&D scientists working on the next generation of in vitro diagnostic applications across the clinical and research settings.
Learn more about the KOD exo(-) DNA Polymerase and KOD Multi Epi DNA Polymerase or explore our KOD One and KOD SYBR qPCR Master Mixes.



Share Article